21. Lesson Conclusion
Lesson Conclusion
ND320 C2 L2 19 Lesson Conclusion
Lesson conclusion
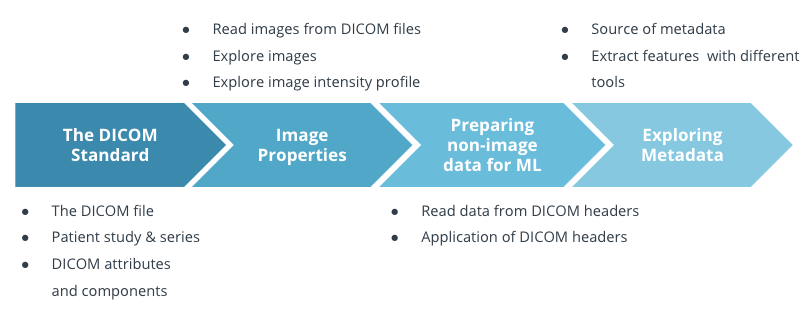
We first talked about the DICOM standard and what a DICOM file is. Then we talked about the patient study and series. After that, we learned that a DICOM file consists of attributes about patient information, patient study, series, and images.
In the second section, we talked about how to read and extract images using the pydicom package, what we should pay attention to when we look at an image, and how to explore image intensity profile at the pixel level.
In the third section, we discussed how to prepare non-image data for ML using the DICOM header. DICOM header has a wealth of information such as patient data, series, and study and we have to explore DICOM header carefully before developing the algorithm.
In the last section, we talked about metadata. Metadata can come from several sources: DICOM header, patient history, and image labels. We need to extract data features before we train the model. Some useful tools are histograms, scatterplots, Pearson correlation coefficient, and co-occurrence matrix.
Further reading
- This website will tell you pretty much everything you ever wanted to know about DICOM
- The github repo for pydicom has lots of documentation, as well as examples of how to use pydicom with DICOM and some sample datasets
- This review article provides a really thorough overview of digital pathology and its history